-Search query
-Search result
Showing 1 - 50 of 491 items for (author: teng & j)

EMDB-35323: 
Cryo-EM structure of the ISFba1 TnpB-reRNA-dsDNA complex
Method: single particle / : Yin M, Zhou F, Zhu Y, Huang Z

PDB-8iaz: 
Cryo-EM structure of the ISFba1 TnpB-reRNA-dsDNA complex
Method: single particle / : Yin M, Zhou F, Zhu Y, Huang Z

EMDB-19562: 
Vimentin intermediate filament protofibril stoichiometry
Method: subtomogram averaging / : Eibauer M, Medalia O

EMDB-19563: 
Vimentin intermediate filament structure (delta tail)
Method: helical / : Eibauer M, Medalia O

EMDB-37847: 
potassium outward rectifier channel SKOR
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

EMDB-37855: 
SKOR D312N L271P double mutation
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

PDB-8wtz: 
potassium outward rectifier channel SKOR
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

PDB-8wui: 
SKOR D312N L271P double mutation
Method: single particle / : Gao X, Sun T, Lu Y, Jia Y, Xu X, Zhang Y, Fu P, Yang G

EMDB-18659: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

EMDB-18660: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

PDB-8quc: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT1
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Pilati N, Marasco A, Gunthorpe M, Alvaro G, Large C, Lakshminaraya B, Williams E, Sauer DB

PDB-8qud: 
Cryo-EM Structure of Human Kv3.1 in Complex with Modulator AUT5
Method: single particle / : Chi G, Mckinley G, Marsden B, Pike ACW, Ye M, Brooke LM, Bakshi S, Lakshminarayana B, Pilati N, Marasco A, Gunthorpe M, Alvaro GS, Large CH, Williams E, Sauer DB

EMDB-16844: 
Vimentin intermediate filament structure
Method: helical / : Eibauer M, Medalia O
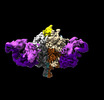
EMDB-41048: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

PDB-8t5c: 
Lassa GPC Trimer in complex with Fab 8.11G and nanobody D5
Method: single particle / : Gorman J, Kwong PD

EMDB-37752: 
Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation
Method: single particle / : Liu M, Wang Y, Su MY, Stjepanovic G

PDB-8wqr: 
Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation
Method: single particle / : Liu M, Wang Y, Su MY, Stjepanovic G

EMDB-37247: 
CULLIN3-KLHL22-RBX1 E3 ligase
Method: single particle / : Su MY

EMDB-41261: 
Cryo-EM structure of Nav1.7 with RLZ
Method: single particle / : Fan X, Huang J, Yan N

EMDB-41262: 
Cryo-EM structure of Nav1.7 with LTG
Method: single particle / : Fan X, Huang J, Yan N
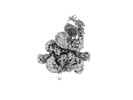
EMDB-37266: 
Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamate dehydrogenase I
Method: single particle / : Su MY

PDB-8w4j: 
Cryo-EM structure of the KLHL22 E3 ligase bound to human glutamate dehydrogenase I
Method: single particle / : Su MY, Su MY

EMDB-37235: 
Human glutamate dehydrogenase I
Method: single particle / : Su MY

EMDB-40352: 
Structure of rat organic anion transporter 1 (OAT1)
Method: single particle / : Dou T, Jiang J

EMDB-40354: 
Structure of rat organic anion transporter 1 (OAT1) in complex with para-aminohippuric acid (PAH)
Method: single particle / : Dou T, Jiang J

EMDB-40355: 
Structure of rat organic anion transporter 1 (OAT1) in complex with probenecid
Method: single particle / : Dou T, Jiang J

EMDB-40948: 
RELION reconstruction of rat organic anion transporter 1 (OAT1)
Method: single particle / : Dou T, Jiang J

PDB-8sdu: 
Structure of rat organic anion transporter 1 (OAT1)
Method: single particle / : Dou T, Jiang J

PDB-8sdy: 
Structure of rat organic anion transporter 1 (OAT1) in complex with para-aminohippuric acid (PAH)
Method: single particle / : Dou T, Jiang J

PDB-8sdz: 
Structure of rat organic anion transporter 1 (OAT1) in complex with probenecid
Method: single particle / : Dou T, Jiang J

EMDB-36668: 
Cryo-EM structure of the zebrafish P2X4 receptor in complex with BX430
Method: single particle / : Hattori M, Shen C

EMDB-36669: 
Cryo-EM structure of the zebrafish P2X4 receptor in complex with BAY-1797
Method: single particle / : Hattori M, Shen C

PDB-8jv5: 
Cryo-EM structure of the zebrafish P2X4 receptor in complex with BX430
Method: single particle / : Hattori M, Shen C

PDB-8jv6: 
Cryo-EM structure of the zebrafish P2X4 receptor in complex with BAY-1797
Method: single particle / : Hattori M, Shen C

EMDB-36751: 
Outward_facing SLC15A4 monomer
Method: single particle / : Zhang SS, Chen XD, Xie M

EMDB-36752: 
Outward-facing SLC15A4 dimer
Method: single particle / : Zhang SS, Chen XD, Xie M

EMDB-36753: 
SLC15A4_TASL complex
Method: single particle / : Zhang SS, Chen XD, Xie M

EMDB-28910: 
Glycan-Base ConC Env Trimer
Method: single particle / : Olia AS, Kwong PD

EMDB-40462: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus pregnenolone sulfate
Method: single particle / : Legesse DH, Fan C, Teng J, Zhuang Y, Howard RJ, Noviello CM, Lindahl E, Hibbs RE

EMDB-40503: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus allopregnanolone
Method: single particle / : Legesse DH, Hibbs RE

EMDB-40506: 
Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus dehydroepiandrosterone sulfate
Method: single particle / : Legesse DH, Hibbs RE
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search












 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
